Note
Go to the end to download the full example code
2.13. Heterogeneous boundary conditions
This example implements a spatially coupled SIR model with the following dynamics for the density of susceptible, infected, and recovered individuals:
\[\begin{split}\partial_t s &= D \nabla^2 s - \beta is \\
\partial_t i &= D \nabla^2 i + \beta is - \gamma i \\
\partial_t r &= D \nabla^2 r + \gamma i\end{split}\]
Here, \(D\) is the diffusivity, \(\beta\) the infection rate, and \(\gamma\) the recovery rate.
0%| | 0/10.0 [00:00<?, ?it/s]
Initializing: 0%| | 0/10.0 [00:00<?, ?it/s]
0%| | 0/10.0 [00:00<?, ?it/s]
2%|2 | 0.23/10.0 [00:00<00:09, 1.00s/it]
7%|7 | 0.71/10.0 [00:00<00:03, 2.57it/s]
29%|##9 | 2.94/10.0 [00:00<00:01, 6.25it/s]
80%|######## | 8.02/10.0 [00:00<00:00, 8.84it/s]
80%|######## | 8.02/10.0 [00:01<00:00, 7.43it/s]
100%|##########| 10.0/10.0 [00:01<00:00, 9.26it/s]
100%|##########| 10.0/10.0 [00:01<00:00, 9.26it/s]
import numpy as np
from pde import CartesianGrid, DiffusionPDE, ScalarField
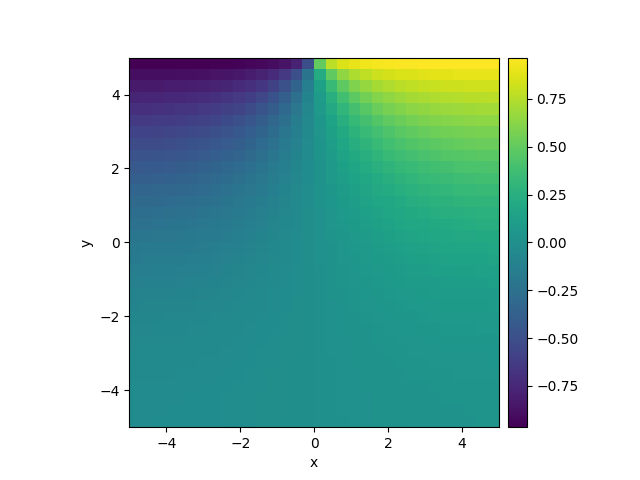
# define grid and an initial state
grid = CartesianGrid([[-5, 5], [-5, 5]], 32)
field = ScalarField(grid)
# define the boundary conditions, which here are calculated from a function
def bc_value(adjacent_value, dx, x, y, t):
"""return boundary value"""
return np.sign(x)
bc_x = "derivative"
bc_y = ["derivative", {"value_expression": bc_value}]
# define and solve a simple diffusion equation
eq = DiffusionPDE(bc=[bc_x, bc_y])
res = eq.solve(field, t_range=10, dt=0.01, backend="numpy")
res.plot()
Total running time of the script: ( 0 minutes 1.261 seconds)