Note
Go to the end to download the full example code
2.20. Custom Class for coupled PDEs
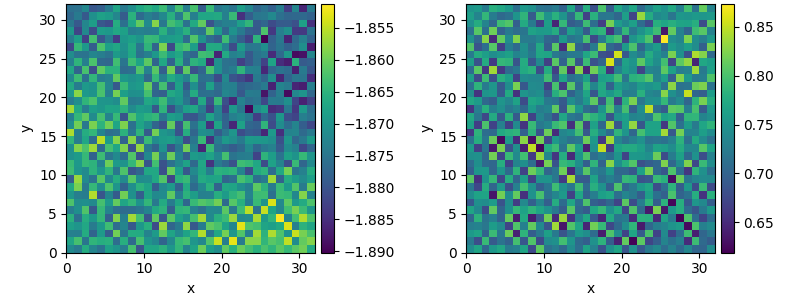
This example shows how to solve a set of coupled PDEs, the spatially coupled FitzHugh–Nagumo model, which is a simple model for the excitable dynamics of coupled Neurons:
\[\begin{split}\partial_t u &= \nabla^2 u + u (u - \alpha) (1 - u) + w \\
\partial_t w &= \epsilon u\end{split}\]
Here, \(\alpha\) denotes the external stimulus and \(\epsilon\) defines the recovery time scale. We implement this as a custom PDE class below.
0%| | 0/100.0 [00:00<?, ?it/s]
Initializing: 0%| | 0/100.0 [00:00<?, ?it/s]
0%| | 0/100.0 [00:00<?, ?it/s]
0%| | 0.24/100.0 [00:00<01:40, 1.00s/it]
1%| | 0.73/100.0 [00:00<00:39, 2.51it/s]
3%|2 | 2.88/100.0 [00:00<00:16, 5.73it/s]
8%|7 | 7.57/100.0 [00:00<00:11, 7.88it/s]
14%|#4 | 14.5/100.0 [00:01<00:09, 8.89it/s]
23%|##2 | 22.97/100.0 [00:02<00:08, 9.37it/s]
32%|###2 | 32.31/100.0 [00:03<00:07, 9.63it/s]
42%|####2 | 42.14/100.0 [00:04<00:05, 9.79it/s]
52%|#####2 | 52.23/100.0 [00:05<00:04, 9.88it/s]
62%|######2 | 62.42/100.0 [00:06<00:03, 9.95it/s]
73%|#######2 | 72.68/100.0 [00:07<00:02, 10.00it/s]
83%|########2 | 82.97/100.0 [00:08<00:01, 10.04it/s]
93%|#########3| 93.27/100.0 [00:09<00:00, 10.07it/s]
93%|#########3| 93.27/100.0 [00:09<00:00, 9.41it/s]
100%|##########| 100.0/100.0 [00:09<00:00, 10.09it/s]
100%|##########| 100.0/100.0 [00:09<00:00, 10.09it/s]
from pde import FieldCollection, PDEBase, UnitGrid
class FitzhughNagumoPDE(PDEBase):
"""FitzHugh–Nagumo model with diffusive coupling"""
def __init__(self, stimulus=0.5, τ=10, a=0, b=0, bc="auto_periodic_neumann"):
super().__init__()
self.bc = bc
self.stimulus = stimulus
self.τ = τ
self.a = a
self.b = b
def evolution_rate(self, state, t=0):
v, w = state # membrane potential and recovery variable
v_t = v.laplace(bc=self.bc) + v - v**3 / 3 - w + self.stimulus
w_t = (v + self.a - self.b * w) / self.τ
return FieldCollection([v_t, w_t])
grid = UnitGrid([32, 32])
state = FieldCollection.scalar_random_uniform(2, grid)
eq = FitzhughNagumoPDE()
result = eq.solve(state, t_range=100, dt=0.01)
result.plot()
Total running time of the script: ( 0 minutes 10.358 seconds)