Note
Go to the end to download the full example code
2.25 Custom PDE class: SIR model
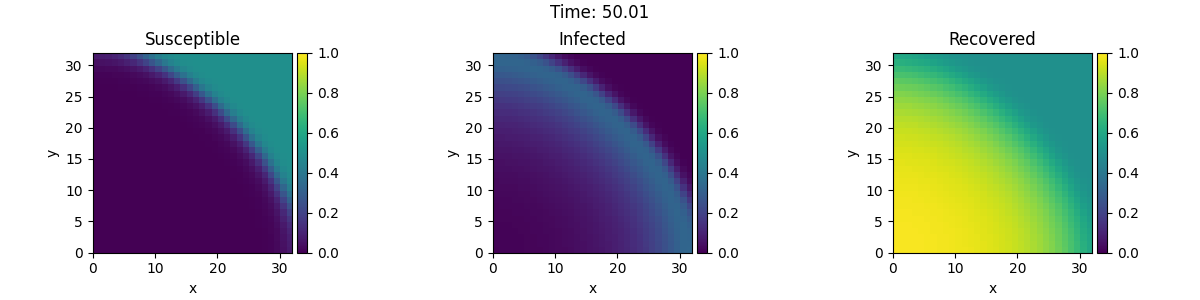
This example implements a spatially coupled SIR model with the following dynamics for the density of susceptible, infected, and recovered individuals:
\[\begin{split}\partial_t s &= D \nabla^2 s - \beta is \\
\partial_t i &= D \nabla^2 i + \beta is - \gamma i \\
\partial_t r &= D \nabla^2 r + \gamma i\end{split}\]
Here, \(D\) is the diffusivity, \(\beta\) the infection rate, and \(\gamma\) the recovery rate.
0%| | 0/50.0 [00:00<?, ?it/s]
Initializing: 0%| | 0/50.0 [00:00<?, ?it/s]
0%| | 0/50.0 [00:00<?, ?it/s]
0%| | 0.02/50.0 [00:00<34:44, 41.70s/it]
0%| | 0.04/50.0 [00:00<17:25, 20.93s/it]
1%| | 0.46/50.0 [00:00<01:36, 1.95s/it]
4%|▍ | 2.14/50.0 [00:01<00:25, 1.89it/s]
11%|█ | 5.61/50.0 [00:01<00:12, 3.46it/s]
21%|██ | 10.58/50.0 [00:02<00:09, 4.14it/s]
31%|███▏ | 15.72/50.0 [00:03<00:07, 4.79it/s]
43%|████▎ | 21.74/50.0 [00:04<00:05, 4.97it/s]
55%|█████▌ | 27.51/50.0 [00:05<00:04, 5.30it/s]
68%|██████▊ | 33.89/50.0 [00:06<00:03, 5.35it/s]
80%|███████▉ | 39.85/50.0 [00:07<00:01, 5.55it/s]
93%|█████████▎| 46.34/50.0 [00:08<00:00, 5.56it/s]
93%|█████████▎| 46.34/50.0 [00:09<00:00, 5.10it/s]
100%|██████████| 50.0/50.0 [00:09<00:00, 5.50it/s]
100%|██████████| 50.0/50.0 [00:09<00:00, 5.50it/s]
from pde import FieldCollection, PDEBase, PlotTracker, ScalarField, UnitGrid
class SIRPDE(PDEBase):
"""SIR-model with diffusive mobility"""
def __init__(
self, beta=0.3, gamma=0.9, diffusivity=0.1, bc="auto_periodic_neumann"
):
super().__init__()
self.beta = beta # transmission rate
self.gamma = gamma # recovery rate
self.diffusivity = diffusivity # spatial mobility
self.bc = bc # boundary condition
def get_state(self, s, i):
"""generate a suitable initial state"""
norm = (s + i).data.max() # maximal density
if norm > 1:
s /= norm
i /= norm
s.label = "Susceptible"
i.label = "Infected"
# create recovered field
r = ScalarField(s.grid, data=1 - s - i, label="Recovered")
return FieldCollection([s, i, r])
def evolution_rate(self, state, t=0):
s, i, r = state
diff = self.diffusivity
ds_dt = diff * s.laplace(self.bc) - self.beta * i * s
di_dt = diff * i.laplace(self.bc) + self.beta * i * s - self.gamma * i
dr_dt = diff * r.laplace(self.bc) + self.gamma * i
return FieldCollection([ds_dt, di_dt, dr_dt])
eq = SIRPDE(beta=2, gamma=0.1)
# initialize state
grid = UnitGrid([32, 32])
s = ScalarField(grid, 1)
i = ScalarField(grid, 0)
i.data[0, 0] = 1
state = eq.get_state(s, i)
# simulate the pde
tracker = PlotTracker(interrupts=10, plot_args={"vmin": 0, "vmax": 1})
sol = eq.solve(state, t_range=50, dt=1e-2, tracker=["progress", tracker])
Total running time of the script: (0 minutes 9.427 seconds)